Genotyping
Short tandem repeat (STR) microsatellite lengths are highly polymorphic in human populations, but their stability during life span of an individual has enabled STR DNA typing as the method of choice for identification. Cell lines are usually populations of cells that are actively evolving with time in culture and with increased passage level in a laboratory. Many cases of misidentified cell lines probably arise as a result of samples being inadvertently mixed-up/switched, mislabeled, or cross-contaminated.
Microsatellite instability (MSI) is the result of a deficient mismatch repair system (MMR) and has been reported first in cases of hereditary non-polyposis colorectal cancer (HNPCC), a cancer-prone familial syndrome. In case of a MSI-positive cell line the MMR system is unable to correct DNA replication errors at STR loci, which are prone to slippage mistakes by replicative DNA polymerases, like STR sequences. Both, mixed as well as MSI-positive cell lines can show additional STR alleles in one or more STR loci. Finally, two fundamentally different causes can be the reason for multiple alleles: Drifted alleles by MSI or the presence of a foreign human cell line.
Diploid allele distribution of MMR proficient cell lines remained stable and unaltered independent of time point of establishment, passage number or grade of nuclear ploidy of a respective line. In contrast, cell lines harboring mutations/deletions within MLH1 or MSH2 as essential members of the MMR system showed the presence of multiple STR alleles at different chromosomal loci. The phenomenon of allelic drift within STRs has a serious impact on authentication of human cell lines, since STR typing serves as a reference technology in cell line authentication at major cell banks ATCC, RIKEN, JCRB and DSMZ.
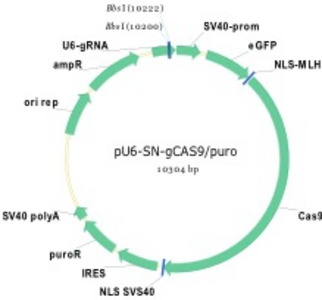
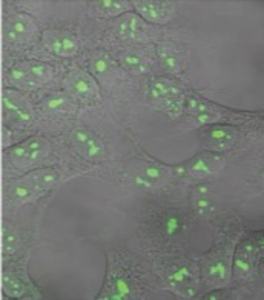
Figure 1) Mediating loss-of-function (Lof) mutations of genes in the human genome
A vector (left) was constructed with a N-terminal fusion of GFP and CAS9 gene in order to visualize gene transfer for detection of recipient cells HT-1080 (right). A U6 promoter is additionally located on the plasmid, which produces the guide RNA simultaneously with expression of CAS9. By designing suitable guide RNAs and cloning, different genes can be knocked out in parallel or sequentially in a very short time.
Project: Revealing the driving forces of Microsatellite Instability
A project for an evaluation of MMR components by gene disruption investigates which genes of the MMR system influence the drift of STR alleles and how high the impact is. To this end, a CRISPR/CAS9 based disruption technique will be used to reveal to what extent genes from the group of DNA helicases, DNA exonucleases and DNA polymerases play a role in the establishment of MSI in addition to the main actors of the MMR system (MLH1, PMS2, MSH2, MSH6). Potential candidates as disruption targets for MSI investigation as a brief summary are as follows:
MutL: MLH1, PMS2, PMS1, MLH3,
MutS: MSH2, MSH6, MSH3,
Helicases: BLM, BRIP,
Nucleases: FAN1, RBBP8, EXO1,
Polymerases: POLH,
DNA repair Platforms: BRCA1, MYC, BLM
The candidate genes should be knocked out both individually and in meaningful combinations and the corresponding individual clones characterized. With the detection of the absence of detectable amounts of protein from the targeted candidate genes, the successful lof mutations will be read out by the drifting of STR alleles into routine STR typings and by the monomorphic MSI markers BAT25 and BAT26. If it is possible to show an impact of specific genes which are involved in the development of MSI, a corresponding analysis should be a pre-condition for new established cell lines, before cells and respective DNA reference profiles are entered into any STR databases of cell line repositories.
Selected References
- Roesner LM, Mielke C, Faehnrich S, Merkhoffer Y, Dittmar KE, Drexler HG, Dirks WG: Localization of MLH3 at the centrosomes. Int J Mol Sci. 2014 Aug 11;15(8):13932-7.
- Roesner LM, Mielke C, Fähnrich S, Merkhoffer Y, Dittmar KE, Drexler HG, Dirks WG: Stable expression of MutLγ in human cells reveals no specific response to mismatched DNA, but distinct recruitment to damage sites. J Cell Biochem. 2013 Oct;114(10):2405-14.
- Dirks WG and Drexler HG: STR DNA typing of human cell lines: detection of intra- and interspecies cross-contamination. Methods Mol Biol. 2013;946:27-38.
- Dirks WG, MacLeod RA, Nakamura Y, Kohara A, Reid Y, Milch H, Drexler HG, Mizusawa H: Cell line cross-contamination initiative: an interactive reference database of STR profiles covering common cancer cell lines. Int J Cancer. 2010 Jan 1;126(1):303-4.