Uncovering the hidden biosynthetic potential of actinomycetes by SARP activation for drug discovery (SARPact)
(Leibniz Cooperative Excellence project K445/2022)
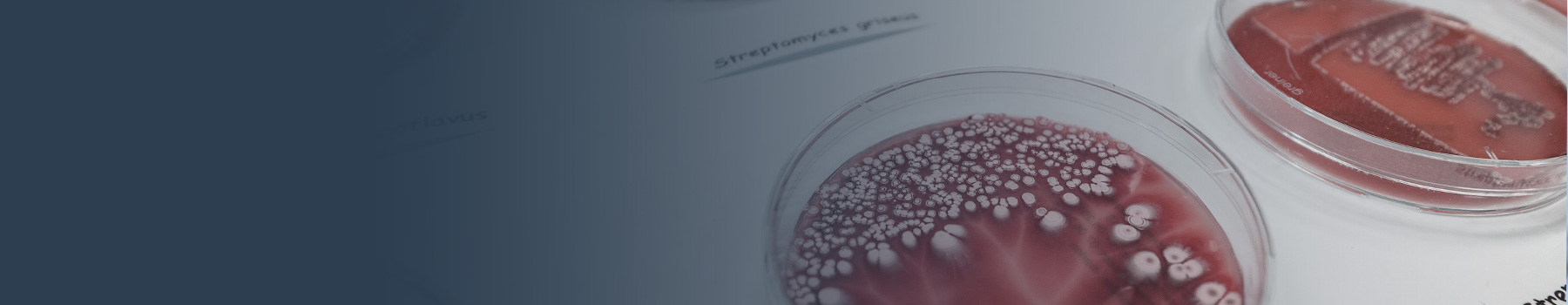
Actinomycetes are the most important source for antibiotics as they account for up to two-thirds of all antibiotics in clinical use today. Based on their genetic setup, these microorganisms can produce many more anti-infectives, but with the current lab methods the production of these "hidden" antibiotic treasures is not activated. Some tools to force such activation exist, but a widely applicable, easy and general method to force the actinomycetes to reveal their secret armory against competitors and infection is missing. With SARPact we will provide such a central tool. SARP regulators are very abundant in actinomycetes, where they act as direct activators of biosynthetic gene clusters, encoding different types of antibiotics. With SARPact we will establish a large-scale targeted process for the activation of silent biosynthetic gene clusters by using SARP-type regulators. This will involve genome mining and genetic engineering of sequenced actinomycetes from the DSMZ strain collection for SARP-guided cluster activation. Engineered strains will be analyzed for novel natural compounds with antimicrobial properties, of which lead substances will be isolated, structurally elucidated and profiled. Concomitantly, the molecular mechanistic principle of the SARPs will be investigated, which will allow for a knowledge-based optimization of the activator. SARPact builds upon the complementary expertise of three academic and one industrial partner, covering the fields of genetic engineering, genome mining, protein structural biology, natural product chemistry, and bioactivity screenings.
Cooperating partners: Leibniz-Institut für Pflanzenbiochemie (IPB); Helmholtz-Zentrum für Infektionsforschung (HZI); BASF SE
Precision Access to Antibiotic Compounds and Targets (PAACT)
(DZIF cooperation project TTU 09.826)
In the collaborative DZIF project PAACT we screen the DSMZ strain collection for potent and interesting natural compound producers. Thereby focus is lead on rare and/or underexplored taxa that are prioritized based on phylogenomic analysis and which display a high genetic potential for secondary metabolite biosynthesis. Furthermore, we concentrate on unique compound classes, such as phosphonate antibiotics. Here, we employ genome mining approaches for the identification of potential phosphonate producers and prove phosphonate production with the help of a phosphonate-specific bioassay.
Generation of novel and resistance-breaking agents by mutasynthesis approaches
(Project BWST_WSF-035 funded by the Baden-Württemberg Stiftung, project ended in 2021)
Antibiotic-resistant pathogens are an increasing threat to human health. Streptogramins are last-resort antibiotics, which are used to treat infections caused by resistant pathogens. In the current project we aim to optimize the streptogramin antibiotic pristinamycin I with a mutasynthesis approach by altering the bioactivity-relevant phenylglycine-residue of the compound. The generated pristinamycin derivatives shall have improved antibiotic activities and resistance-breaking characteristics. The research project is supported by the Baden-Württemberg Stiftung and is carried out as a collaborative study together with AG Stegmann (University of Tübingen) and Jung-Won Youn/AG Sprenger (University of Stuttgart).
Exploiting underexplored translation inhibitors and combinations thereof
(DZIF cooperation project TTU 09.819, project ended in 2020)
The bacterial ribosome is a hot spot for the action of many successful antibiotics. However, not all promising binding sites at the ribosome are therapeutically exploited. The aim of the project is to identify and characterize new and/or underexplored protein synthesis inhibitors. Here we develop a genome sequence-based screening approach for the identification of potential producer strains of protein synthesis inhibitors. The screening strategy is applied to strains of the Tübingen and the DSMZ actinomycetes strain collection. Protein synthesis inhibitor activity from samples of the identified strains is analyzed in vitro transcription/translation assays, as well as in a mode of action-specific reporter system in collaboration with AG Brötz-Oesterhelt (University of Tübingen).
Isolation of new unique natural compounds from unknown actinomycetes (NAbaUnAk)
(BMBF-funded project 16GW0124K, project ended in 2019)
Actinomycetes are the most versatile antibiotic producers as they synthesize two-thirds of all known natural antibiotics. Sampling of unique biotopes turned out to be an efficient way to isolate unknown actinomycetes that potentially produce novel natural compounds. Indonesia is one of the most species-rich countries in the world. This biodiversity might also be reflected by a microbial species diversity. Thus, especially Indonesian soils should serve as an excellent source for unknown actinomycetes strains that may produce novel antimicrobial compounds. Our aim is to isolate novel natural compounds from Indonesian actinomycetes and investigate their potential as new anti-infective drug leads.
Optimisation of the lysolipins, potent broad-spectrum antibiotics from Streptomyces
(DZIF project TTU 09.912; project ended in 2019)
Lysolipin is an extraordinarily potent antibacterial natural product, with a minimum inhibitory concentration (MIC) in the low nanomolar range against Gram-positive organisms and a higher, but still nanomolar MIC against Gram-negative microorganisms. Its mechanism of action is not yet fully understood. However, its usability as antibacterial agent is limited due to the fact that is also shows cytotoxicity. First experiments have shown that structural modifications of the lysolipin core structure can differentially influence antibacterial activity and cytotoxicity. The aim of the project was to develop lysolipin derivatives with improved bioactivities by genetic engineering approaches.
For previous projects please visit: uni-tuebingen.de/de/30400